JASON 3.1 brings substantial new features for Diffusion Ordered SpectroscopY (DOSY). Reporting the results now includes a table of diffusion coefficients and charts showing the fit of individual signal decays.

Figure 1 A DOSY spectrum (top left), results table (top right) and signal decay of the TMS peak (bottom)
Let us look into the parameter options inside the NMR processing function of DOSY. I am going to summarize them systematically including all options, old and new parameters alike.
Improved interface
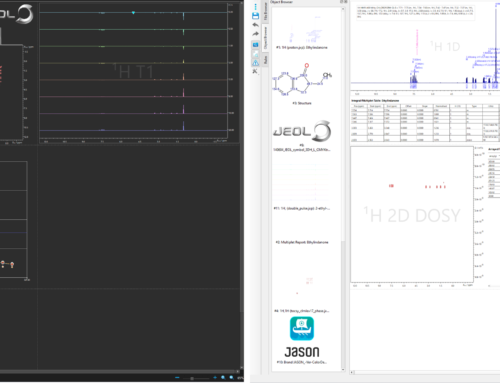
The graphical widget has been redesigned following user feedback. The default, basic mode, shows only those parameters which we most often wish to customize from sample to sample. We can select how to generate data for the fitting process, using integrals by default, peak heights, or all data points of the spectrum.

Figure 2 The processing panel in JASON 3.1 showing basic (left) and expert (right) mode in the DOSY widget
The other three options only impact the visualization of the DOSY plot and do not affect the values of the resulting diffusion coefficients. The “Lowest D” and “Highest D” values specify the range of diffusion coefficients to be shown in the resulting DOSY plot. The input simply takes the values using scientific notation and are given in units of m2/s. In Fig.1, this corresponds to 0.8×10-10 and 0.5×10-8 covering a wide range of typical diffusion coefficients for small molecules in solution. It is desirable to narrow the region of interest to improve digitization without the need for a very large number of points in the diffusion dimension. For wide ranges, increase the ‘Digitization’ parameter. Projections constructed from DOSY plots can be sensitive to under-digitization.
We can switch the interface to the “Expert mode” to access all parameters. On the top of the widget, the same parameters are controlled as in basic mode. This is followed by three further groups of parameters including options for the sequence, gradient and diffusion delay parameters. Manual override of the default values is usually not needed, unless JASON fails to recognize the experiment and extract the parameters correctly. We aim to provide automatic identification for any of the default sequences from supported vendors. Please contact us (support@JEOLjason.com) if you notice problems. The exception to this is the gradient calibration factor (T/m factor), which may need adjustments for accurate diffusion results.
List of parameters
Here we provide the list of all parameters implemented in JASON 3.1, including obsolete parameters. They are given in their respective order in JASON processing template files (.jjp).
- “Points” is used to control the number of points to use from the gradient list of arrayed data. The introduction of better support to ‘drop-points’ made this parameter obsolete, and it is no longer active from version 3.1. We still save it in processing templates to provide compatibility with JASON 3.0 and older versions.
- “Lowest D” sets the slowest diffusion coefficient to plot in the 2D DOSY. Note, this is not exactly the value of the last data point due to impact of digital resolution
- “Highest D” sets the fastest diffusion coefficient to plot in the 2D DOSY.
- “Array list” is the name of the key under which the gradient list is internally stored in Jason_parameters. It is automatically set by JASON when we open data. Usually it is array(g), difflist and gzlvl1 for JEOL/Delta, Bruker/TopSpin and Varian data files, respectively. Users only need to change this if a customized pulse program implementation is not recognized correctly or the data format is from unsupported Vendor. ‡
- “T/m factor” is the calibration factor to convert gradient list elements to actual amplitudes for DOSY. JASON automatically sets this parameter for nominal values as defined in original vendor parameters. Accurate DOSY measurements require calibration of the spectrometer (specific to each NMR probe used) once with a known sample, typically 1% H2O in D2
- “γ [rad s-1 T-1]” is the gyromagnetic ratio. By default, it is set for the nuclide in the relevant dimension. This defaults to 1H if the nuclide is not defined (see nuclides in jason_parameters).
- “Δ [s]” is the nominal diffusion time. From version 3.1, the pulse sequence specific corrected diffusion time is automatically calculated and changes to this parameter are only required if the pulse program was not recognized by JASON.
- “Pulse” is the duration of the diffusion-encoding gradient pulse. Our definition for bipolar gradients follows the same notation as given by Davy Sinnaeve in Concepts in Magnetic Resonance, 40A, 39-65 (2012). JASON automatically imports this correctly for standard DOSY experiments.
- “Digitisation” is the number of points in the diffusion domain for constructing the DOSY plots. This has no relation to the number of elements in the gradient array list. Very large values (>4000) will consume more memory, disk space and slow down GUI responses. Consider ‘zooming’ into the relevant diffusion region for plotting higher resolution DOSY. Please note, both horizontal and vertical projections are sensitive to under-digitization of the diffusion domain.
- “Use” allows selection between using integrals, peak heights or all data points. The use of integrals is preferred because it is the most comprehensively developed framework. When using peak heights the peak positions are set automatically. When using integrals, the stacked view of the raw DOSY data helps to visualize integral regions and they can be set without any restrictions. The “all points” option is recommended to be used with Shrink (or Resample) applied at the end of the direct dimension part of the processing list to ‘cut out’ a smaller region of interest; leaving approximately 1k points only. As each point will be fitted individually, the processing can become excessively slow when using very large number of points in this mode.
- “Shape” menu provides selection of the gradient shape used. This is relevant not only for the correct magnitude calculation of the diffusion encoding but also for the automatic correction of the effective diffusion time. Currently, the supported shapes are rectangular, half-sine or user defined. The user-defined option allows the user to take into account any shape, but it is not compatible with automatic calculations of the effective diffusion time. In such cases, provide both the relevant factor (next parameter) for the gradient shape, but also specify manually the effective diffusion time.
- “shape factor” is the multiplication factor for non-rectangular gradient pulse shapes, reflecting their effective loss of encoding with respect to the rectangular case.
- “Use shape factor” is a flag to take into account the previous parameter as a multiplier when reading the gradient list values. Note, there is a reason for this seemingly unnecessary flag to provide correct processing for all vendors. Some vendors provide the gradient list values already corrected by the shape factor and others not. Even when gradient list is already corrected, we still need the correct setting of the shape factor for the automatic calculation of the effective diffusion time. JASON automatically set these parameters for default experiments‡
Introduced from JASON version 3.1
- “Sequence” menu allows the user to select the type of DOSY pulse sequence that was used to collect to the data. This is only relevant for the automatic calculation of the effective diffusion time. Current options are bipolar, monopolar, oneShot, Bipolar_CC, Monopolar_CC, and Unknown. The ’ _CC’ stands for convection compensated. The “Unknown” identifier is a placeholder for other variants, which may become supported in later JASON versions.
- “InterPulseDelay [s]” is the time between the two bipolar gradient pulses. Typically, the sum of the gradient stabilization delay, the observe 180 pulse duration, and any additional hardware delays for transmitter or gradient gating.‡ This parameter is only visible for bipolar type sequences (Bipolar, oneShot or Bipolar_CC)
- “Alpha” is the unbalancing factor used in oneShot type sequences. Also referred to as ‘kappa’ in some vendor parameters. This parameter is only visible for the oneShot type sequences (consequently not shown in the example in Fig. 2)
- “Δ’ [s]” is the corrected value of the diffusion time. It is set automatically when next parameter is checked, otherwise the user can define it manually.
- “Auto” checkbox controls the automatic calculation of the previous parameter value, the corrected diffusion time.
- “Basic mode” or “Expert mode” switch the interface to show only a few or all parameters when the DOSY processing widget is expanded.
‡ Please let us know (support@JEOLjason.com) if you find you need to change this parameter manually to help us improve our automatic data interpretation for all of our users.